-
Laurent Modolo authoredLaurent Modolo authored
- Introduction
- Data set : nycflights13
- Data type
- filter rows
- Logical operators
- Missing values
- Challenges
- Arrange rows with arrange()
- Missing values
- Challenges
- Select columns with select()
- Helper functions
- Challenges
- Add new variables with mutate()
- mutate()
- Useful creation functions
- See you in R#5: Pipping and grouping
- To go further: Data transformation and color sets.
- RColorBrewer & Ghibli
- Viridis
- Volcano Plot
title: "R.4: data transformation"
author: "Laurent Modolo [laurent.modolo@ens-lyon.fr](mailto:laurent.modolo@ens-lyon.fr), Hélène Polvèche [hpolveche@istem.fr](mailto:hpolveche@istem.fr)"
date: "2021"
output:
rmdformats::downcute:
self_contain: true
use_bookdown: true
default_style: "dark"
lightbox: true
css: "http://perso.ens-lyon.fr/laurent.modolo/R/src/style.css"rm(list=ls())
knitr::opts_chunk$set(echo = TRUE)
knitr::opts_chunk$set(comment = NA)klippy::klippy(
position = c('top', 'right'),
color = "white",
tooltip_message = 'Click to copy',
tooltip_success = 'Copied !')Introduction
The goal of this practical is to practice data transformation with tidyverse.
The objectives of this session will be to:
- Filter rows with
filter() - Arrange rows with
arrange() - Select columns with
select() - Add new variables with
mutate()
Solution
install.packages("nycflights13")library("tidyverse")
library("nycflights13")Data set : nycflights13
nycflights13::flightsContains all 336,776 flights that departed from New York City in 2013.
The data comes from the US Bureau of Transportation Statistics, and is documented in ?flights
flightsData type
In programming languages, all variables are not equal.
When you display a tibble you can see the type of a column.
Here is a list of common variable types that you will encounter
- int stands for integers.
- dbl stands for doubles or real numbers.
- chr stands for character vectors or strings.
- dttm stands for date-times (a date + a time).
-
lgl stands for logical, vectors that contain only
TRUEorFALSE. - fctr stands for factors, which R uses to represent categorical variables with fixed possible values.
- date stands for dates.
You cannot add an int to a chr, but you can add an int to a dbl the results will be a dbl.
filter rows
Variable types are important to keep in mind for comparisons.
The filter() function allows you to subset observations based on their values.
What is the results of the following filter command ?
filter(flights, month == 1, day == 1)dplyr functions never modify their inputs, so if you want to save the result, you’ll need to use the assignment operator, <-
Solution
jan1 <- filter(flights, month == 1, day == 1)(dec25 <- filter(flights, month == 12, day == 25))Logical operators
Multiple arguments to filter() are combined with AND: every expression must be TRUE in order for a row to be included in the output.
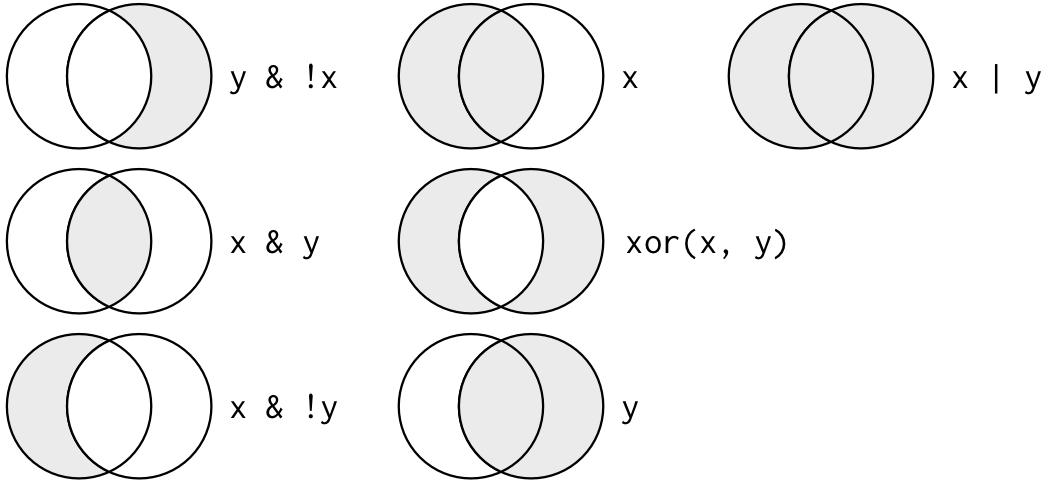
In R you can use the symbols &, |, ! and the function xor() to build other kinds of tests.
filter(flights, month == 11 | month == 12)filter(flights, month %in% c(11, 12))filter(flights, !(arr_delay > 120 | dep_delay > 120))filter(flights, arr_delay <= 120, dep_delay <= 120)Combinations of logical operators is a powerful programmatic way to select subset of data. Keep in mind, however, that long logical expression can be hard to read and understand, so it may be easier to apply successive small filters instead of one long one.
Missing values
One important feature of R that can make comparison tricky is missing values, or NAs for Not Availables.
Indeed each of the variable type can contain either a value of this type (i.e., 2 for an int) or nothing.
The nothing recorded in a variable status is represented with the NA symbol.
As operations with NA values don t make sense, if you have NA somewhere in your operation, the results will be NA
NA > 5
10 == NA
NA + 10However, you can test for NAs with the function is.na():
is.na(NA)filter() only includes rows where the condition is TRUE; it excludes both FALSE and NA values. If you want to preserve missing values, ask for them explicitly:
df <- tibble(x = c(1, NA, 3))
filter(df, x > 1)
filter(df, is.na(x) | x > 1)Challenges
Solution
filter(flights, arr_delay >= 60 | arr_delay <= 120)filter(flights, dest %in% c("IAH", "HOU"))Solution
filter(flights, is.na(dep_time))Solution
NA ^ 0 # ^ 0 is always 1 it's an arbitrary rule not a computation
NA | TRUE # if a member of a OR operation is TRUE the results is TRUE
FALSE & NA # if a member of a AN operation is FALSE the results is TRUE
NA * 0 # here we have a true computation
Arrange rows with arrange()
arrange() works similarly to filter() except that instead of selecting rows, it changes their order.
arrange(flights, year, month, day)Solution
arrange(flights, desc(dep_delay))Missing values
Missing values are always sorted at the end:
arrange(tibble(x = c(5, 2, NA)), x)
arrange(tibble(x = c(5, 2, NA)), desc(x))Challenges
- Find the most delayed flight.
- Find the flight that left earliest.
- How could you arrange all missing values to the start ?
Solution
Find the most delayed flight.
arrange(flights, desc(dep_delay))Find the flight that left earliest.
arrange(flights, dep_delay)How could you arrange all missing values to the start
arrange(tibble(x = c(5, 2, NA)), desc(is.na(x)))
Select columns with select()
select() allows you to rapidly zoom in on a useful subset using operations based on the names of the variables.
You can select by column names
select(flights, year, month, day)By defining a range of columns
select(flights, year:day)Or you can do a negative (-) to remove columns.
select(flights, -(year:day))Helper functions
here are a number of helper functions you can use within select():
-
starts_with("abc"): matches names that begin with"abc". -
ends_with("xyz"): matches names that end with"xyz". -
contains("ijk"): matches names that contain"ijk". -
num_range("x", 1:3): matchesx1,x2andx3.
See ?select for more details.
Challenges
- Brainstorm as many ways as possible to select
dep_time,dep_delay,arr_time, andarr_delayfromflights.
Solution
select(flights, contains("time") | contains("delay"))
select(flights, contains("_") & !starts_with("sched") & !starts_with("time"))vars <- c("year", "month", "day", "dep_delay", "arr_delay")Solution
select(flights, one_of(vars))- Does the result of running the following code surprise you? How do the select helpers deal with case by default? How can you change that default?
select(flights, contains("TIME"))Solution
select(flights, contains("TIME", ignore.case = FALSE))
Add new variables with mutate()
It’s often useful to add new columns that are functions of existing columns. That’s the job of mutate().
Solution
(flights_sml <- select(flights, year:day, ends_with("delay"), distance, air_time))
mutate()
mutate(tbl, new_var_a = opperation_a, ..., new_var_n = opperation_n)mutate() allows you to add new columns (new_var_a, ... , new_var_n) and to fill them with the results of an operation.
We can create a gain column to check if the pilot managed to compensate is departure delay
mutate(flights_sml, gain = dep_delay - arr_delay)Solution
flights_sml <- mutate(flights_sml,
gain = dep_delay - arr_delay,
speed = distance / air_time * 60
)Solution
mutate(
flights,
dep_time = (dep_time %/% 100) * 60 +
dep_time %% 100,
sched_dep_time = (sched_dep_time %/% 100) * 60 +
sched_dep_time %% 100
)Useful creation functions
- Offsets:
lead()andlag()allow you to refer to leading or lagging values. This allows you to compute running differences (e.g.x - lag(x)) or find when values change (x != lag(x)). - Cumulative and rolling aggregates: R provides functions for running sums, products, mins and maxes:
cumsum(),cumprod(),cummin(),cummax(); and dplyr providescummean()for cumulative means. - Logical comparisons,
<,<=,>,>=,!=, and== - Ranking: there are a number of ranking functions, but you should start with
min_rank(). There is alsorow_number(),dense_rank(),percent_rank(),cume_dist(),ntile()
See you in R#5: Pipping and grouping
To go further: Data transformation and color sets.
There are a number of color palettes available in R, thanks to different packages such as RColorBrewer, Viridis or Ghibli.
We will use them here to decorate our graphs, either on data already studied in the training, mpg, or on more specialized data such as lists of differentially expressed genes ( GSE86356 )
install.packages(c("ghibli", "RColorBrewer", "viridis"))library(tidyverse)
library(RColorBrewer)
library(ghibli)
library(viridis)RColorBrewer & Ghibli
Using mpg and the 'ggplot2' package, reproduce the graph studied in session 2, 3.1: color mapping.
Modify the colors representing the class of cars with the palettes Dark2 of RColorBrewer, then MononokeMedium from Ghibli.
ggplot(data = mpg, mapping = aes(x = displ, y = hwy, color = class)) +
geom_point()Go to the links to find the appropriate function: they are very similar between the two packages.
Solution
ggplot(data = mpg, mapping = aes(x = displ, y = hwy, color = class)) +
geom_point() +
scale_color_brewer(palette = "Dark2")ggplot(data = mpg, mapping = aes(x = displ, y = hwy, color = class)) +
geom_point() +
scale_colour_ghibli_d("MononokeMedium")The choice of colors is very important for the comprehension of a graphic. Some palettes are not suitable for everyone. For example, for people with color blindness, color gradients from green to red, or from yellow to blue should be avoided.
To display only Brewer palettes that are colorblind friendly, specify the option colorblindFriendly = TRUE as follows:
display.brewer.all(colorblindFriendly = TRUE)Viridis
viridis package provide a series of color maps that are designed to improve graph readability for readers with common forms of color blindness and/or color vision deficiency.
For the next part, we will use a real data set. Anterior tibial muscle tissue was collected from 20 patients, with or without confirmed myotonic dystrophy type 1 (DM1). Illumina RNAseq was performed on these samples and the sequencing data are available on GEO with the identifier GSE86356.
First, we will use the gene count table of these samples, formatted for use in ggplot2 ( pivot_longer() function ).
Open the csv file using the read_csv2() function. The file is located at "http://perso.ens-lyon.fr/laurent.modolo/R/session_4/Expression_matrice_pivot_longer_DEGs_GSE86356.csv".
Solution
expr_DM1 <- read_csv2("http://perso.ens-lyon.fr/laurent.modolo/R/session_4/Expression_matrice_pivot_longer_DEGs_GSE86356.csv")
expr_DM1With this tibble, use ggplot2 and the geom_tile() function to make a heatmap.
Fit the samples on the x-axis and the genes on the y-axis.
Tip: Transform the counts into log10(x + 1) for a better visualization.
Solution
ggplot(expr_DM1, aes(samples, Genes, fill= log1p(counts))) +
geom_tile() +
labs(y="Genes", x = "Samples") +
theme(
axis.text.y = element_text(size= 4),
axis.text.x = element_text(size = 4, angle = 90)
)Nota bene : The elements of the axes, and the theme in general, are modified in the theme() function.
With the default color gradient, even with the transformation, the heatmap is difficult to study.
R interprets a large number of colors, indicated in RGB, hexadimal, or just by name. For example :
{width=400px}With scale_fill_gradient2() function, change the colors of the gradient, taking white for the minimum value and 'springgreen4' for the maximum value.
Solution
ggplot(expr_DM1, aes(samples, Genes, fill= log1p(counts))) +
geom_tile() +
scale_fill_gradient2(low = "white", high = "springgreen4") +
labs(y="Genes", x = "Samples") +
theme(
axis.text.y = element_text(size= 4),
axis.text.x = element_text(size = 4, angle = 90)
)It s better, but still not perfect! Now let s use the viridis color gradient for this graph.
Solution
ggplot(expr_DM1, aes(samples, Genes, fill= log1p(counts))) +
geom_tile() +
scale_fill_viridis_c() +
labs(y="Genes", x = "Samples") +
theme(
axis.text.y = element_text(size= 4),
axis.text.x = element_text(size = 4, angle = 90)
)Volcano Plot
For this last exercise, we will use the results of the differential gene expression analysis between DM1 vs WT conditions.
Open the csv file using the read_csv2() function. The file is located at "http://perso.ens-lyon.fr/laurent.modolo/R/session_4/EWang_Tibialis_DEGs_GRCH37-87_GSE86356.csv".
Solution
tab <- read_csv2("http://perso.ens-lyon.fr/laurent.modolo/R/session_4/EWang_Tibialis_DEGs_GRCH37-87_GSE86356.csv")
tabTo make a Volcano plot, displaying different information about the significativity of the variation thanks to the colors, we will have to make a series of modifications on this table.
With mutate() and ifelse() fonctions, we will have to create :
-
a column 'sig' : it indicates if the gene is significant ( TRUE or FALSE ). Thresholds : baseMean > 20 and padj < 0.05 and abs(log2FoldChange) >= 1.5
-
a column 'UpDown' : it indicates if the gene is Up regulated or Down regulated.
Solution
tab.sig <- tab %>%
mutate(sig = baseMean > 20 & padj < 0.05 & abs(log2FoldChange) >= 1.5 ) %>%
mutate(UpDown = ifelse(
baseMean > 20 & padj < 0.05 & log2FoldChange >= 1.5,
"Up",
ifelse(
baseMean > 20 & padj < 0.05 & log2FoldChange <= -1.5,
"Down",
"NO"
)))
tab.sigWe want to see the top10 DEGs on the graph. For this, we will use the package ggrepel.
Install and load the ggrepl package.
Solution
install.packages("ggrepel")library(ggrepel)Let s filter our table into a new variable, top10, to keep only the top 10 according to the adjusted pvalue. The smaller the adjusted pvalue, the more significant.
Tips : You can use the function slice_min()
Solution
top10 <- tab.sig %>%
filter(sig == TRUE) %>%
slice_min(n = 10, padj)The data is ready to be used to make a volcano plot!
Tips 1 : Don t forget the transformation of the adjusted pvalue.
Tips 2 : Feel free to search your favorite Web browser for help.
Tips 3 : geom_label_repel() function needs a new parameter 'data' and 'label' in aes parameters.
ggplot(tab.sig, aes(x = log2FoldChange, y = -log10(padj), color = UpDown)) +
geom_point() +
scale_color_manual(values=c("steelblue", "lightgrey", "firebrick" )) +
geom_hline(yintercept=-log10(0.05), col="black") +
geom_vline(xintercept=c(-1.5, 1.5), col="black") +
theme_minimal() +
theme(
legend.position="none"
) +
labs(y="-log10(p-value)", x = "log2(FoldChange)") +
geom_label_repel(data = top10, mapping = aes(label = gene_symbol))
Solution
ggplot(tab.sig, aes(x = log2FoldChange, y = -log10(padj), color = UpDown)) +
geom_point() +
scale_color_manual(values=c("steelblue", "lightgrey", "firebrick" )) +
geom_hline(yintercept=-log10(0.05), col="black") +
geom_vline(xintercept=c(-1.5, 1.5), col="black") +
theme_minimal() +
theme(
legend.position="none"
) +
labs(y="-log10(p-value)", x = "log2(FoldChange)") +
geom_label_repel(data = top10, mapping = aes(label = gene_symbol))